The important package has a succinct interface for obtaining estimates of predictor importance with tidymodels objects. A few of the main features:
There are also recipe steps for supervised feature selection:
step_predictors_retain() can filter the predictors
using a single conditional statement (e.g., absolute correlation with
the outcome > 0.75, etc).step_predictors_best() can retain the most important
predictors for the outcome using a single scoring function.step_predictors_desirability() retains the most
important predictors for the outcome using multiple scoring functions,
blended using desirability functions.The latter two steps can be tuned over the proportion of predictors to be retained.
You can install the development version of important from GitHub with:
install.packages("devtools")
# or
pak::pak("tidymodels/important")The main reason for making important is censored regression models. tidymodels released tools for fitting and qualifying models that have censored outcomes. This included some dynamic performance metrics that were evaluated at different time points. This was a substantial change for us, and it would have been even more challenging to add to other packages.
Let’s look at an analysis that models food delivery times. The outcome is the time between an order being placed and the delivery (all data are complete - there is no censoring). We model this in terms of the order day/time, the distance to the restaurant, and which items are contained in the order. Exploratory data analysis shows several nonlinear trends in the data and some interactions between these trends.
We’ll load the tidymodels and important packages to get started.
The data are split into training, validation, and testing sets.
data(deliveries, package = "modeldata")
set.seed(991)
delivery_split <- initial_validation_split(deliveries, prop = c(0.6, 0.2), strata = time_to_delivery)
delivery_train <- training(delivery_split)The model uses a recipe with spline terms for the hour and distances. The nonlinear trend over the time of order changes on the day, so we added interactions between these two sets of terms. Finally, a simple linear regression model is used for estimation:
delivery_rec <-
recipe(time_to_delivery ~ ., data = delivery_train) |>
step_dummy(all_factor_predictors()) |>
step_zv(all_predictors()) |>
step_spline_natural(hour, distance, deg_free = 10) |>
step_interact(~ starts_with("hour_"):starts_with("day_"))
lm_wflow <- workflow(delivery_rec, linear_reg())
lm_fit <- fit(lm_wflow, delivery_train)First, let’s capture the effect of the individual model terms. These terms are from the derived features in the models, such as dummy variables, spline terms, interaction columns, etc.
set.seed(382)
lm_deriv_imp <-
importance_perm(
lm_fit,
data = delivery_train,
metrics = metric_set(mae, rsq),
times = 50,
type = "derived"
)
lm_deriv_imp
#> # A tibble: 226 × 6
#> .metric predictor n mean std_err importance
#> <chr> <chr> <int> <dbl> <dbl> <dbl>
#> 1 rsq distance_10 50 0.528 0.00655 80.5
#> 2 mae day_Sat 50 1.09 0.0150 72.2
#> 3 mae distance_10 50 2.20 0.0323 68.2
#> 4 mae day_Fri 50 0.877 0.0140 62.7
#> 5 mae day_Thu 50 0.638 0.0130 49.0
#> 6 mae distance_09 50 0.740 0.0156 47.3
#> 7 mae hour_08 50 0.520 0.0136 38.3
#> 8 rsq day_Sat 50 0.118 0.00327 36.2
#> 9 rsq hour_06_x_day_Sat 50 0.146 0.00410 35.7
#> 10 mae hour_08_x_day_Sat 50 0.604 0.0173 34.9
#> # ℹ 216 more rowsUsing mean absolute error as the metric of interest, the top 5 features are:
lm_deriv_imp |>
filter(.metric == "mae") |>
slice_max(importance, n = 5)
#> # A tibble: 5 × 6
#> .metric predictor n mean std_err importance
#> <chr> <chr> <int> <dbl> <dbl> <dbl>
#> 1 mae day_Sat 50 1.09 0.0150 72.2
#> 2 mae distance_10 50 2.20 0.0323 68.2
#> 3 mae day_Fri 50 0.877 0.0140 62.7
#> 4 mae day_Thu 50 0.638 0.0130 49.0
#> 5 mae distance_09 50 0.740 0.0156 47.3Two notes:
The importance scores are the ratio of the mean change in performance and the associated standard error. The mean value is always increasing with importance, no matter which direction is preferred for the specific metric(s).
We can run these in parallel by loading the future package and
specifying a parallel backend using the plan()
function.
There is a plot method that can help visualize the results:
autoplot(lm_deriv_imp, top = 50)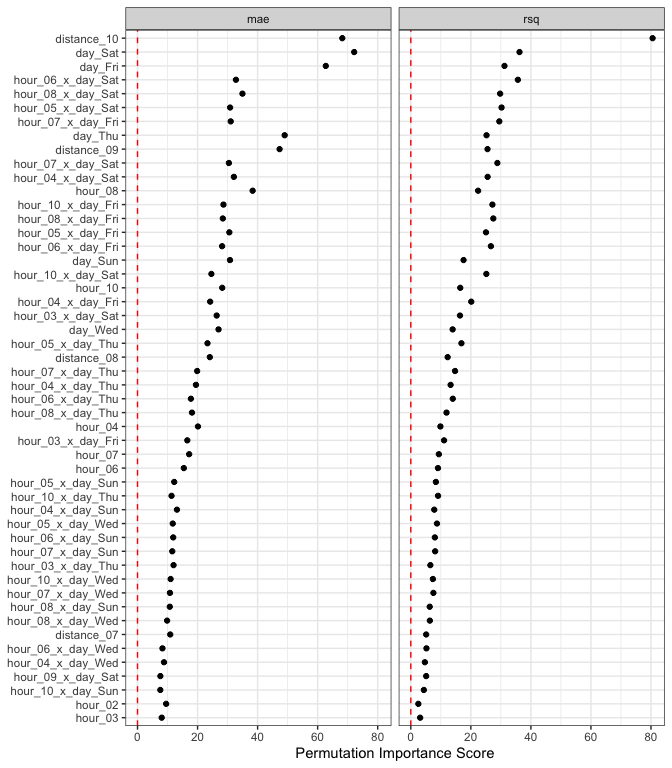
Since there are spline terms and interactions for the hour column, we
might not care about the importance of a term such as
hour_06 (the sixth spline feature). In aggregate, we might
want to know the effect of the original predictor columns. The
type option is used for this purpose:
set.seed(382)
lm_orig_imp <-
importance_perm(
lm_fit,
data = delivery_train,
metrics = metric_set(mae, rsq),
times = 50,
type = "original"
)
# Top five:
lm_orig_imp |>
filter(.metric == "mae") |>
slice_max(importance, n = 5)
#> # A tibble: 5 × 6
#> .metric predictor n mean std_err importance
#> <chr> <chr> <int> <dbl> <dbl> <dbl>
#> 1 mae hour 50 4.10 0.0320 128.
#> 2 mae day 50 1.91 0.0214 89.1
#> 3 mae distance 50 1.46 0.0222 66.0
#> 4 mae item_24 50 0.0489 0.0119 4.11
#> 5 mae item_03 50 0.0337 0.0114 2.95autoplot(lm_orig_imp)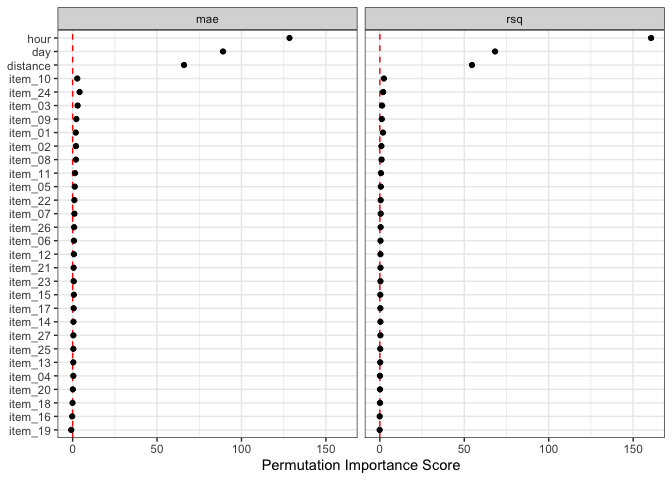
Using the same dataset, let’s illustrate the most common tool for filtering predictors: using random forest importance scores.
important can use any of the “scoring functions” from the filtro package. You can supply one, and the proportion of the predictors to retain:
set.seed(491)
selection_rec <-
recipe(time_to_delivery ~ ., data = delivery_train) |>
step_predictor_best(all_predictors(), score = "imp_rf", prop_terms = 1/4) |>
step_dummy(all_factor_predictors()) |>
step_zv(all_predictors()) |>
step_spline_natural(any_of(c("hour", "distance")), deg_free = 10) |>
step_interact(~ starts_with("hour_"):starts_with("day_")) |>
prep()
selection_rec
#>
#> ── Recipe ──────────────────────────────────────────────────────────────────────
#>
#> ── Inputs
#> Number of variables by role
#> outcome: 1
#> predictor: 30
#>
#> ── Training information
#> Training data contained 6004 data points and no incomplete rows.
#>
#> ── Operations
#> • Feature selection via `imp_rf` on: item_03 item_04, ... | Trained
#> • Dummy variables from: day | Trained
#> • Zero variance filter removed: <none> | Trained
#> • Natural spline expansion: hour distance | Trained
#> • Interactions with: hour_01:day_Tue hour_01:day_Wed, ... | TrainedA list of possible scores is contained in the help page for the recipe steps.
Note that we changed selectors in step_spline_natural()
to use any_of() instead of specific names. Any step
downstream of any filtering steps should be generalized so that there is
no failure if the columns were removed. Using any_of()
selects these two columns if they still remain in the data.
Which were removed?
selection_res <-
tidy(selection_rec, number = 1) |>
arrange(desc(score))
selection_res
#> # A tibble: 30 × 4
#> terms removed score id
#> <chr> <lgl> <dbl> <chr>
#> 1 hour FALSE 48.5 predictor_best_rCIMa
#> 2 day FALSE 13.5 predictor_best_rCIMa
#> 3 distance FALSE 13.3 predictor_best_rCIMa
#> 4 item_10 FALSE 1.18 predictor_best_rCIMa
#> 5 item_01 FALSE 1.01 predictor_best_rCIMa
#> 6 item_24 FALSE 0.160 predictor_best_rCIMa
#> 7 item_02 FALSE 0.0676 predictor_best_rCIMa
#> 8 item_26 TRUE 0.0666 predictor_best_rCIMa
#> 9 item_03 TRUE 0.0593 predictor_best_rCIMa
#> 10 item_22 TRUE 0.0565 predictor_best_rCIMa
#> # ℹ 20 more rows
mean(selection_res$removed)
#> [1] 0.7666667This example shows the basic usage of the recipe. In practice, we would probably do things differently:
prop_terms = tune() in the step and using one of
the tuning functions to find a good proportion.Inappropriate use of these selection steps occurs when it is used before the data are split or outside of a resampling step.
Please note that the important project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.